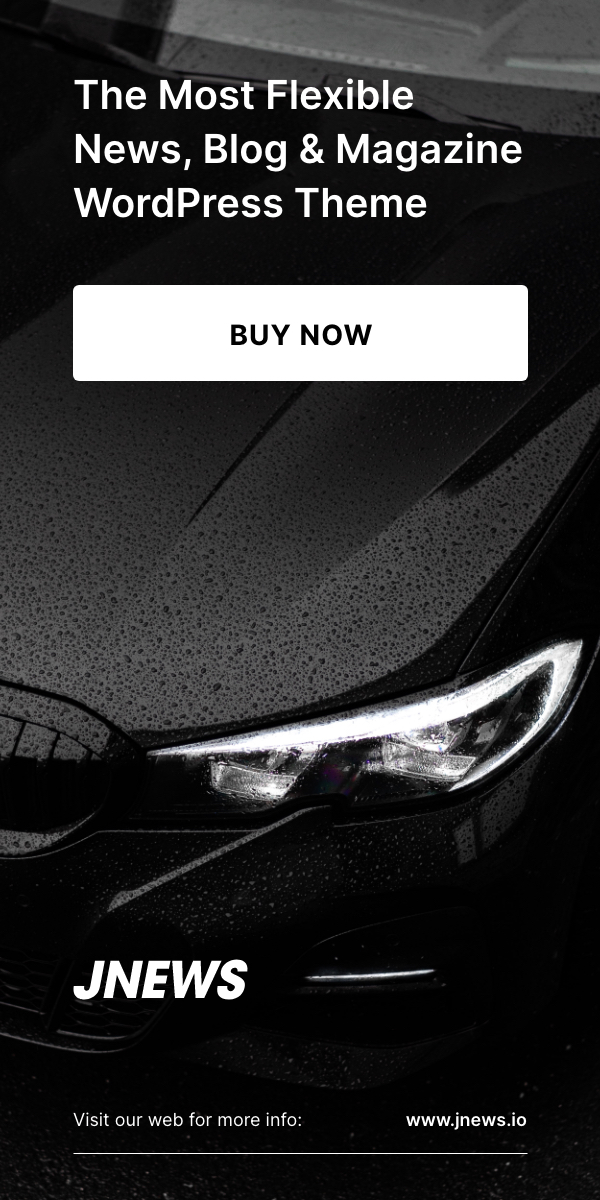
DNA origami nanostructures (blue) can be utilized to program the form of virus particles (gray). The native capsid with a diameter of 28 nanometers is proven in green-grey. Credit score: Mauri A. Kostiainen/Aalto College
Proteins that encapsulate viruses may be molded into outlined shapes utilizing DNA and RNA origami nanostructures.
Bioengineers have found a technique to customise the dimensions and form of virus particles. This new method, which entails merging viral protein constructing blocks and DNA templates, provides potential functions within the fields of vaccine creation and drug supply.
Utilizing Virus Capsid Proteins
Virus capsid proteins, the protecting defend for a virus’s genome, can function a base for creating meticulously structured protein assemblies. Nevertheless, their shapes and geometry primarily depend upon the virus pressure. Reprogramming these assemblies, no matter the unique viral blueprint, presents a tantalizing chance in areas corresponding to drug supply and vaccine growth.
The scientific staff addressed this problem by producing a “structured genome” template for the meeting of capsid proteins. They utilized inflexible DNA origami buildings to forestall deformation of the versatile genome and the formation of undesirable shapes. These buildings are tiny in dimension, starting from tens to lots of of nanometers, however solely product of DNA, which is exactly folded into the specified template form.
The Position of Electrostatic Interactions
“Our method is predicated on electrostatic interactions between the adverse cost of the DNA nanostructures and a positively charged area of the capsid proteins, paired with intrinsic interactions between the only proteins. By altering the quantity of protein used, we are able to fine-tune the variety of highly-ordered protein layers, which encapsulate the DNA origami,” says Iris Seitz, lead creator and doctoral researcher at Aalto College.
“Through the use of DNA origami as a template, we are able to direct the capsid proteins right into a user-defined dimension and form, leading to assemblies that are well-defined, each in size and diameter. By testing quite a lot of DNA origami buildings, we additionally realized how the templates’ geometry affected the entire meeting,” Seitz provides.
Cryogenic Electron Microscopy Imaging
“With the assistance of cryogenic electron microscopy imaging, we had been capable of visualize the extremely ordered proteins upon meeting and, with that, measure even small adjustments within the geometry of the meeting arising from totally different templates,” explains professor Juha Huiskonen, a collaborating scientist from the College of Helsinki.
Relevance and Purposes
“We’ve discovered a easy however efficient technique to (re)direct capsid proteins to a desired form. Our method is adaptable and due to this fact not restricted to a single capsid protein sort, as we demonstrated with capsid proteins from 4 totally different viruses. Moreover, we are able to tweak our template to be extra application-relevant, as an illustration by integrating RNA into the origami, which may subsequently be translated into helpful or site-specific proteins,” explains Aalto professor Mauri Kostiainen, chief of the analysis mission.
Though DNA origami buildings are a promising materials for interfacing organic programs, they endure from instability, particularly within the presence of DNA-degrading enzymes.
In experiments, nevertheless, “we are able to clearly observe that the protein layer effectively protects the encapsulated DNA nanostructures from degradation. By combining safety with the purposeful properties of nucleic acid origami, together with the likelihood to ship DNA or messenger RNA along with different cargo molecules, we consider that our method offers fascinating future instructions for biomedical engineering,” concludes Kostiainen.
Reference: “DNA-origami-directed virus capsid polymorphism” by Iris Seitz, Sharon Saarinen, Esa-Pekka Kumpula, Donna McNeale, Eduardo Anaya-Plaza, Vili Lampinen, Vesa P. Hytönen, Frank Sainsbury, Jeroen J. L. M. Cornelissen, Veikko Linko, Juha T. Huiskonen and Mauri A. Kostiainen, 17 July 2023, Nature Nanotechnology.
DOI: 10.1038/s41565-023-01443-x
This work was performed collectively at Aalto College (Finland) with researchers from the College of Helsinki (Finland), Griffith College (Australia), Tampere College (Finland) and College of Twente (The Netherlands).