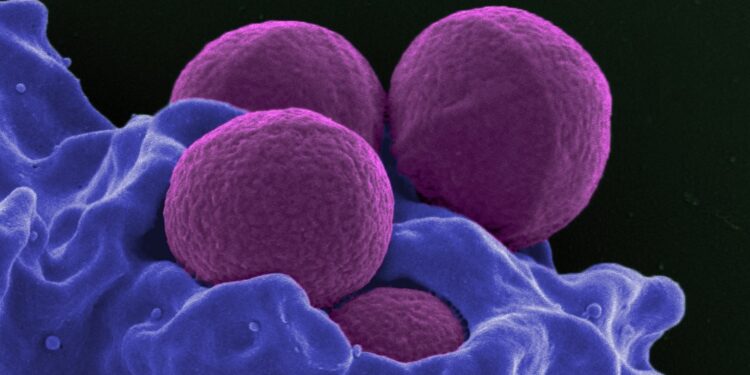
Antibiotics have saved numerous lives and are an important instrument in trendy medication. However we’re dropping floor in our battle towards micro organism. In the midst of the final century, scientists discovered whole new classes of antibiotics. Since then, the tempo of discovery has slowed to a trickle, and the prevalence of antibiotic-resistant micro organism has grown.
There are doubtless antibiotics but to be found, however the chemical universe is just too massive for anybody to go looking. Lately, scientists have turned to AI. Machine studying algorithms can whittle huge numbers of potential chemical configurations all the way down to a handful of promising candidates for testing.
To this point, scientists have used AI to search out single compounds with antibiotic properties. However in a brand new research, published yesterday in Nature, MIT researchers say they’ve constructed and examined a system that may establish complete new courses of antibiotics and predict that are doubtless secure for folks.
The AI sifted over 12 million compounds and located an undiscovered class of antibiotics that proved efficient in mice towards methicillin-resistant Staphylococcus aureus (MRSA), a lethal pressure of drug-resistant bug.
Whereas these AI-discovered antibiotics nonetheless have to show themselves secure and efficient in people by passing the usual gauntlet of scientific testing, the staff believes their work can pace discovery on the entrance finish and, hopefully, enhance our general hit fee.
Exploring Drug House
Scientists are more and more utilizing AI sidekicks to hurry up the method of discovery. Most well-known, maybe, is DeepMind’s AlphaFold, a machine studying program that may mannequin the shapes of proteins, our physique’s primary constructing blocks. The thought is that AlphaFold and its descendants can pace up the arduous strategy of drug analysis. So sturdy is their conviction, DeepMind spun out a subsidiary in 2021, Isomorphic Labs, devoted to doing simply that.
Different AI approaches have additionally proven promise. An MIT group, particularly, has been centered on creating fully new antibiotics to battle superbugs. Their first research, printed in 2020, established the method may work, after they discovered halicin, a beforehand undiscovered antibiotic that could readily take out drug-resistant E. coli.
In a followup earlier this 12 months, the staff took purpose at Acinetobacter baumannii, “public enemy No. 1 for multidrug-resistant bacterial infections,” based on McMaster College’s Jonathan Stokes, a senior creator on the research.
“Acinetobacter can survive on hospital doorknobs and tools for lengthy durations of time, and it may possibly take up antibiotic resistance genes from its surroundings. It’s actually frequent now to search out A. baumannii isolates which can be resistant to almost each antibiotic,” Stokes said at the time.
After combing via 6,680 compounds in simply two hours, the AI highlighted a couple of hundred promising candidates. The staff examined 240 of those that have been structurally completely different from present antibiotics. They surfaced 9 promising candidates, together with one, abaucin, that was fairly efficient towards A. baumannii.
Each research confirmed the method may work, however solely yielded single candidates with no info on why they have been efficient. Machine studying algorithms are, notoriously, black packing containers—what occurs “between the ears” so to talk is usually an entire thriller.
Within the newest research, the group took purpose at one other recognized adversary, MRSA, solely this time they chained a number of algorithms collectively to enhance outcomes and higher illuminate the AI’s reasoning.
Flipping the Swap
The staff’s newest antibiotic bloodhound skilled on some 39,000 compounds, together with their chemical construction and talent to kill MRSA. Additionally they skilled separate fashions to foretell the toxicity of a given compound to human cells.
“You may characterize principally any molecule as a chemical construction, and in addition you inform the mannequin if that chemical construction is antibacterial or not,” Felix Wong, a postdoc at IMES and the Broad Institute of MIT and Harvard, told MIT News. “The mannequin is skilled on many examples like this. For those who then give it any new molecule, a brand new association of atoms and bonds, it may possibly inform you a likelihood that that compound is predicted to be antibacterial.”
As soon as full, the staff fed over 12 million compounds into the system. The AI narrowed this huge listing all the way down to round 3,600 compounds organized into 5 courses—primarily based on their constructions—it predicted would have some exercise towards MRSA and be minimally poisonous to human cells. The staff settled on a last listing of 283 candidates for testing.
Of those, they discovered two from the identical class—that’s, they’d comparable structural elements believed to contribute to antimicrobial exercise—that have been fairly efficient. In mice, the antibiotics fought each a pores and skin an infection and a systemic an infection by taking out 90 % of MRSA micro organism current.
Notably, whereas their earlier work tackled Gram-negative micro organism by disrupting cell membranes, MRSA is Gram-positive and has thicker partitions.
“We’ve got fairly sturdy proof that this new structural class is lively towards Gram-positive pathogens by selectively dissipating the proton driving force in micro organism,” Wong stated. “The molecules are attacking bacterial cell membranes selectively, in a method that doesn’t incur substantial harm in human cell membranes.”
By making their AI explainable, the staff hopes to zero in on constructions which may inform future searches or contribute to the design of more practical antibiotics within the lab.
Ultimate Exams
The important thing factor to notice right here is that though it seems the brand new antibiotics have been efficient in mice on a really small scale, there’s a protracted technique to go earlier than you’d be prescribed one.
New medication bear rigorous testing and scientific trials, and lots of, even promising candidates, don’t make it via to the opposite aspect. The sphere of AI-assisted drug discovery, extra usually, is still in the early stages in this respect. The primary AI-designed drugs are now in clinical trials, however none have but been authorised.
Nonetheless, the hope is to extra rapidly inventory the pipeline with higher candidates.
It will possibly take three to 6 years to find a brand new antibiotic appropriate for scientific trials, according to the College of Pennsylvania’s César de la Fuente, whose lab is doing comparable work. Then you could have the trials themselves. With antibiotic resistance on the rise, we could not have that form of time, to not point out the actual fact antibiotics don’t have the return on funding different medication do. Any assistance is welcome.
“Now, with machines, we’ve been in a position to speed up [the timeline],” de la Fuente told Scientific American. “In my and my colleagues’ personal work, for instance, we are able to uncover in a matter of hours 1000’s or a whole lot of 1000’s of preclinical candidates as a substitute of getting to attend three to 6 years. I feel AI basically has enabled that.”
It’s early but, but when AI-discovered antibiotics show themselves worthy within the coming years, maybe we are able to preserve the higher hand in our long-standing battle towards micro organism.
Picture Credit score: A human white blood cell ingesting MRSA (purple) / National Institute of Allergy and Infectious Diseases, National Institutes of Health